
6 Protein–protein interactions can be subsequently elucidated by observing the fluorescence signals from the restructured fluorescent proteins under a fluorescence microscope a few days after transformation. 4,5 Expression cassettes of the two fusion proteins can be transiently introduced into plant cells through conventional plant transformation. 1–4 In protein–protein interaction studies using BiFC, two proteins thought to potentially interact with each other are fused to two nonfluorescent fragments of a fluorescent protein. The bimolecular fluorescence complementation (BiFC) assay is one powerful tool for visualizing interactions between two proteins during a biological process. How these interacting biomolecules participate in specific biological reactions is intriguing, because protein interactions are highly complex and dynamic. Introduction Plant cells create functional proteins that synergistically interact with each other to function in particular metabolic processes and in cellular mechanism. We present a cutting-edge technique for codelivery of multiple biomolecules into plant cells that can be used for elucidation of functional correlations and for metabolic engineering. We demonstrate that multiple biomolecule/CPP cargos can be simultaneously internalized by a particular plant cell, albeit with different efficiencies. Here, we employed the engineered CPP KH 9-BP100 as a carrier to deliver multiple biomolecules into plant cells and performed a bimolecular fluorescence complementation assay to assess the simultaneous introduction of multiple biomolecules. Cell-penetrating peptides (CPPs) have demonstrated remarkable abilities to introduce diverse biomolecules into various plant species. However, the currently existing biomolecule delivery methods face difficulties in delivering multiple components into plant cells, mediating transgene expression, and maintaining the stability of the numerous components and lead to delays in biomolecular function. Exogenous codelivery of multiple biomolecules is an essential step for elucidation of the biological significance of these molecules and enables various biotechnological applications in plants. The Tukey procedure explained above is valid only with equal sample sizes for each treatment level.Plant cells contain groups of biomolecules that participate together in a particular biological process.
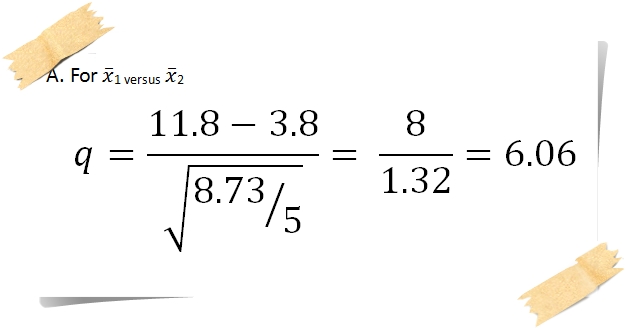
These letters can be added to figures summarizing the results of the ANOVA. We need to add a factor of "b" to show this: 29.20Ĭontinuing down the line, we now calculate the next difference: \(28.60 - 21.00=7.60\), exceeding the critical \(w\), so we now add a "c": 29.20Īgain, we need to go back and check to see if the third-largest also differs from the smallest: \(25.87 - 21.00=4.87\), which it does. Here \(28.6 - 25.87 = 2.73\), less than the critical \(w\) of 2.824, so these two means do not differ significantly. This is a step that sets up a back-and-forth process. Now we have to consider whether or not the second-largest and third-largest differ significantly. Then calculate the difference between the largest and third-largest means, \(29.20 - 25.87=3.33\), which exceeds the critical \(w\) of 2.824, so we can label these with a "b" to show this difference is significant: 29.20

Start with the largest and second-largest means and calculate the difference, \(29.20 - 28.60 = 0.60\), which is less than our \(w\) of 2.824, so we indicate there is no significant difference between these two means by placing the letter "a" under each: 29.20 Step 2: Rank the means, calculate differencesįor the greenhouse example, we rank the means as: 29.20


 0 kommentar(er)
0 kommentar(er)
